Though I am getting a little annoyed at the breathless prose about how it’s “like photosynthesis” and might be a way to sustain astronauts during long space flights and so on.
The story’s about a fungus they found growing (thriving, even) inside the reactor at Chernobyl, despite all the radiation the fungus is exposed to in there.
What the original paper – which you can find here from PubMed Central (and where you can find what the study actually shows, rather than the somewhat lower-content hype found in most news reports on it) – seems to show based on my hasty undergraduate-level reading is that the fungi do grow faster when exposed to “ionizing radiation”, and that it appears to be due to melanin in the plant getting energy from the radiation (and passing that energy on to the fungus to use for growth).
This is actually pretty spiffy, but really – so far – doesn’t look like “photosynthesis” at all. They’re not testing for any kind of carbon fixation, and I’m guessing that if there is any carbon fixation going on, that it doesn’t generate oxygen in the process. It also seems unlikely to me that even then, the fungus can grow autotrophically. This would seem to drastically reduce the possibility of this stuff ever being Purina® Astronaut Chow – you’d still need some other way to get the carbon dioxide out of the Astronaut’s air and put oxygen back in it. If you’re going to do that, you might as well just use plants (or cyanobacteria) and eat THEM.
Still, the implication that you could adapt some melanin-producing fungus to absorb “radiation” and turn it into useful materials of some kind is spiffy, even if it’s not going to allow us to turn nuclear fission plants and spent nuclear fuel depots into fungus-powered anti-global-warming-gas powerhouses.
One thing’s bugging me, though. I obviously don’t have enough understanding of how “ionizing radiation” behaves at a biochemical level, since I’m wondering if it’s proper for everyone to treat “radiation” (both from flying electrons and from high-energy light) as some sort of generic substance, whose only useful attribute is how much energy it has.
As far as I know, most of the “radiation” that the fungus inside the Chernobyl reactor is getting is Gamma-radiation – basically high-energy light (one step above “X-rays”, two steps above sunburn-causing Ultraviolet light). What the researchers are hitting their test-subjects with looks like it’s mainly “Beta”-radiation (which is to say – electrons)*. In both cases it’s “ionizing” radiation, which is to say (more or less) that the radiation knocks electrons off of atoms that it runs into in both cases, and in the ideal “spherical horse” world of a Physicist, the same amount of energy is going to knock the same amount of electrons loose from various molecules and therefore have the same effect, right?
Except I’m having trouble convincing myself that’s a valid assumption here. The results seem to show that exposure to radiation is somehow resulting in the melanin in the fungus being able to “reduce” a chemical (changing “NAD+” into “NADH”) that can potentially in turn dump electrons into the beginning of the Electron Transport Chain to in turn provide biological energy in the form of ATP…
Can one reasonably assume that the mechanism by which this happens would be the same regardless of the form of ionizing radiation? The big deal with melanin seems to be that it absorbs a wide range of light wavelengths (which is why it looks black to dark-brown, and why it protects skin from Ultraviolet radiation…) which implies that absorbing the gamma radiation is where the energy is coming from that makes the fungus thrive in the Chernobyl reactor building. I guess I’m just having trouble picturing how a much more massive, slower-moving electron could have precisely the same effect as a virtually massless, much faster photon. (Yes, I know that beta and gamma radiation are said to have the same amount of “effect” on living tissue per unit of energy…)
Is it possible that the melanin is directly “capturing” the beta particles (electrons), while gamma radiation is kicking electrons off of something ELSE, and melanin is then only indirectly taking up those? For that matter, is it possible that in both cases it’s just something silly like the radiation inducing hydrolysis of water, and it’s just hydrogen gas supplying the reducing power? Thinking about this is making me feel dumb – can anyone reading this explain what I’m missing here?…
I suppose I could just cheat and ask someone in the biology department. We’ve GOT a professor who ought to know – her research has specifically focussed on zapping prokaryotes with “ionizing radiation” (electrons from the college’s linear accelerator)…But that would rob my dear readers of the chance to participate here…
* – okay, it’s probably even more complicated than that. If I understand what the paper is describing and what my Minister Of Funky Physics Knowledge showed me, the source of the “ionizing radiation” for the experiments is radioactive Tungsten(W) and Rhenium (Re) (A “188Re/188W Isotope Generator”). W-188 gives off beta particles when it decays to Re-188. But Re-188 can go through some sort of funky subatomic rearrangement before it decays so that it can EITHER give off beta particles OR gamma-rays as it decays down to stable Osmium-188. I have no idea what the proportion between beta and gamma is at that step (the “conversion efficiency”) so it’s possible there’s enough gamma radiation coming out to do something, regardless of what the beta particles are doing. (The experiment doesn’t do any comparisons with “pure” gamma radiation, which I imagine is not simple to arrange…). So now I’m even MORE confused. Thanks, physics. Thanks a lot.
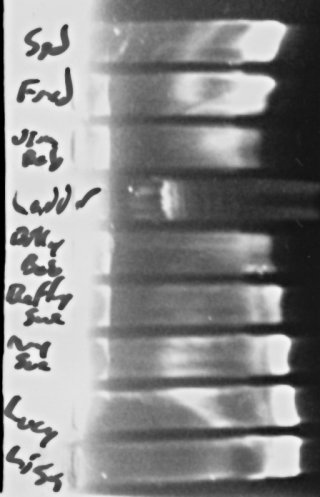 To the left, there, you can see the results. Interestingly, if you look just to the right of where the “wells” are in the gel, in several lanes you can make out a visible band of obviously large DNA molecules, which I presume are genomic DNA from the bacteria. The fact that they seem brighter in the lanes where the 16s band at the end (or at least, I’m ASSUMING that’s what that is, and my reaction worked) is dimmer tends to support my suspicion that I’m “swamping” my reaction with the template DNA and making it hard for the reaction to work sometimes.
To the left, there, you can see the results. Interestingly, if you look just to the right of where the “wells” are in the gel, in several lanes you can make out a visible band of obviously large DNA molecules, which I presume are genomic DNA from the bacteria. The fact that they seem brighter in the lanes where the 16s band at the end (or at least, I’m ASSUMING that’s what that is, and my reaction worked) is dimmer tends to support my suspicion that I’m “swamping” my reaction with the template DNA and making it hard for the reaction to work sometimes.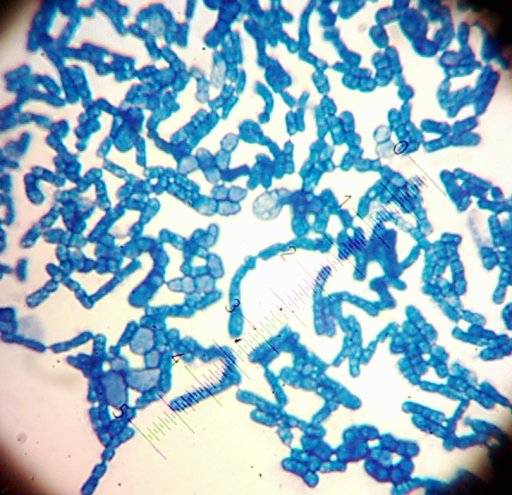
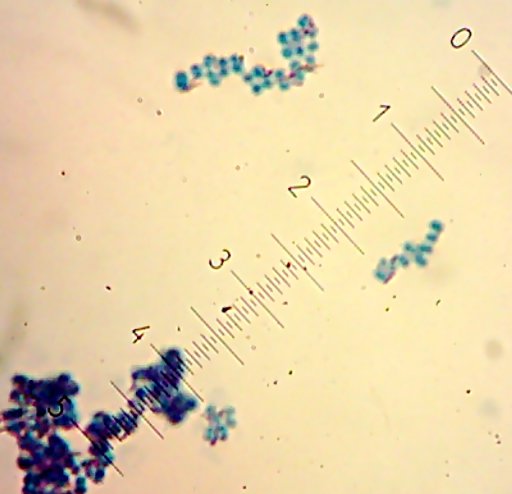
 I also got two more Coccoid-Cluster-type Gram-positive bacterial isolates. The look pretty much the same under the microscope, though one had gooey wet, slightly larger colonies than the other’s smaller, hard-lump colonies. I see another one of those
I also got two more Coccoid-Cluster-type Gram-positive bacterial isolates. The look pretty much the same under the microscope, though one had gooey wet, slightly larger colonies than the other’s smaller, hard-lump colonies. I see another one of those 
![Sid the [lacto?]bacillus-type-thing](http://www.bigroom.org/images/Sid.jpg)
