If you’ve got a culture of a single type of bacteria and you want to identify it, the standard method is to figure out the sequence of one particular gene, the 16s rDNA gene. That is – it’s the gene which encodes the a piece of RNA that gets used by the ribosome in part of the process of “reading” which amino acids to link together to make a particular protein. This is something that every prokaryote known has, and parts of it are conserved, so they’re similar enough to compare, while other parts can vary a lot, providing enough “difference” to tell different organisms apart.
To figure out the sequence, you use PCR to “amplify” this particular gene, making lots of copies of it so that the sequencing machine can clearly see the signal from each part of the sequence. And before you can do that, you have to get the DNA out of the cell relatively intact.
That part can be a pain. There are lots of different ways people have come up with (and made special canned “kits” out of) – you can use chemicals to try to dissolve the cells and let all their guts (including the DNA) out, you can try to mash them up with tiny glass beads in a “bead-beating” machine, you can stick them in a blender, you can even just boil them for a while…then usually you go through several steps of centrifuging and mixing with different chemicals and then centrifuging again until you’ve hopefully finally got the DNA out and gotten rid of most of the other cell bits. And, hopefully, you haven’t accidentally chopped up the DNA too much to use in the process.
Fortunately, there’s a trick you can sometimes use, referred to as “Colony PCR”. In it, you literally just touch the top of your colony of cells and shake them off directly into the PCR tube. Then you just include an extra 5-10 minutes of 95°C heating to hopefully cook open enough of the cells to release DNA (and cook the cell’s enzymes to death so they don’t degrade the DNA and interfere with the PCR).
Not real reliable if you’re trying to do anything quantitative, like trying to figure out how many copies of a gene are in each cell, or trying to get an accurate estimate of how many cells of one type or another are in a mixed culture, but if you just need as much of a particular bit of DNA as you can get – such as for sequencing – a lot of people use this.
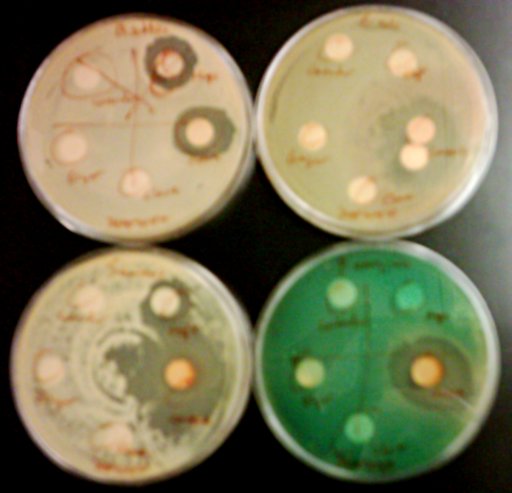
I just tried it on my Lambic isolates. Two of the 8 bacterial cultures worked beautifully. I’m pretty sure the problem with the other 6 was just the sheer amount of bacteria I ended up adding to the reaction – too much seems to “swamp” the PCR process and keep it from working. I’ll try it again this week. But it does seem to indicate that it works, at least.
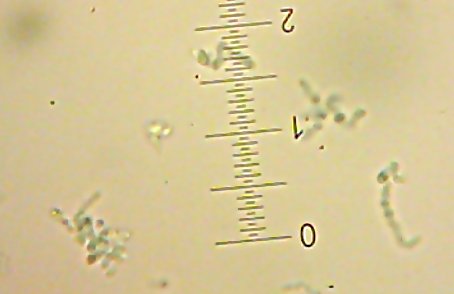
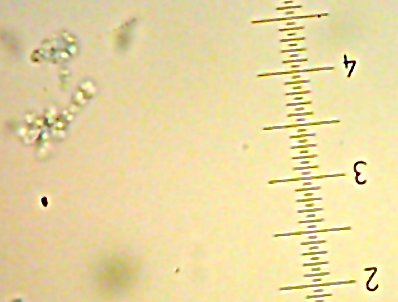
![Sid the [lacto?]bacillus-type-thing](http://www.bigroom.org/images/Sid.jpg)